Mission & Objectives
Advancing fundamental understanding of dopaminergic and kisspeptin systems in puberty and metabolic regulation.
Our Mission
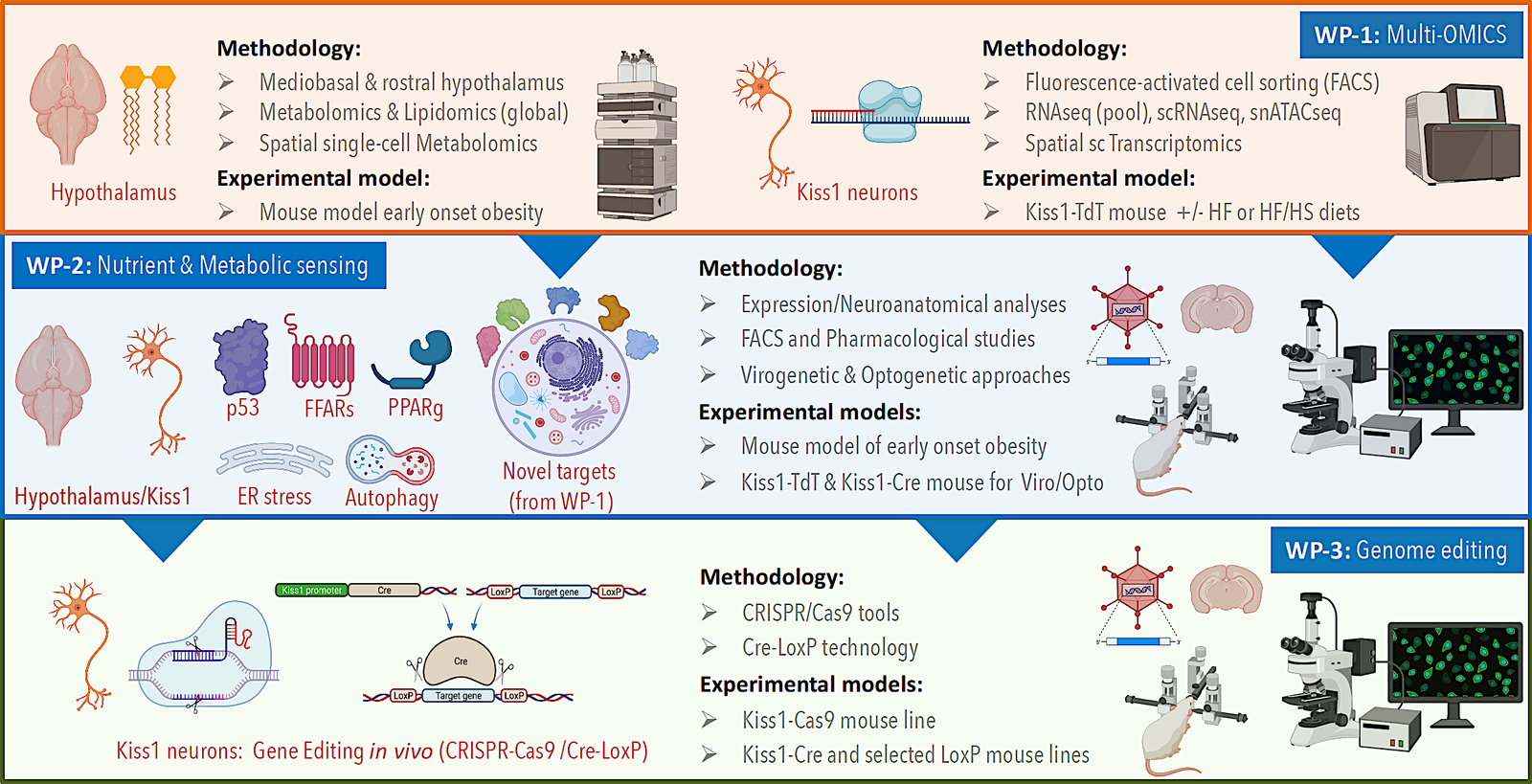
DOPA-Kiss aims to generate a comprehensive framework for understanding how dopaminergic and kisspeptin neuronal networks integrate metabolic and hormonal cues to regulate the onset of puberty and maintain reproductive health. By combining advanced multi-omics, functional neurobiology, and genetic manipulation approaches, the project seeks to identify key molecular pathways and their roles in linking nutritional status to reproductive function.Scientific Rationale
The timing of puberty is a critical determinant of lifelong health, with both early and delayed onset associated with increased risk of metabolic, reproductive, and psychological disorders. Despite its importance, the precise neural and molecular mechanisms that integrate metabolic information into the central control of puberty remain poorly understood.
DOPA-Kiss addresses this gap by focusing on two key neuronal systems—dopaminergic and kisspeptin—that are positioned at the interface of metabolism and reproduction, aiming to uncover their individual and combined contributions to the regulation of pubertal timing.
Core Objectives
The project is structured around three interlinked objectives:

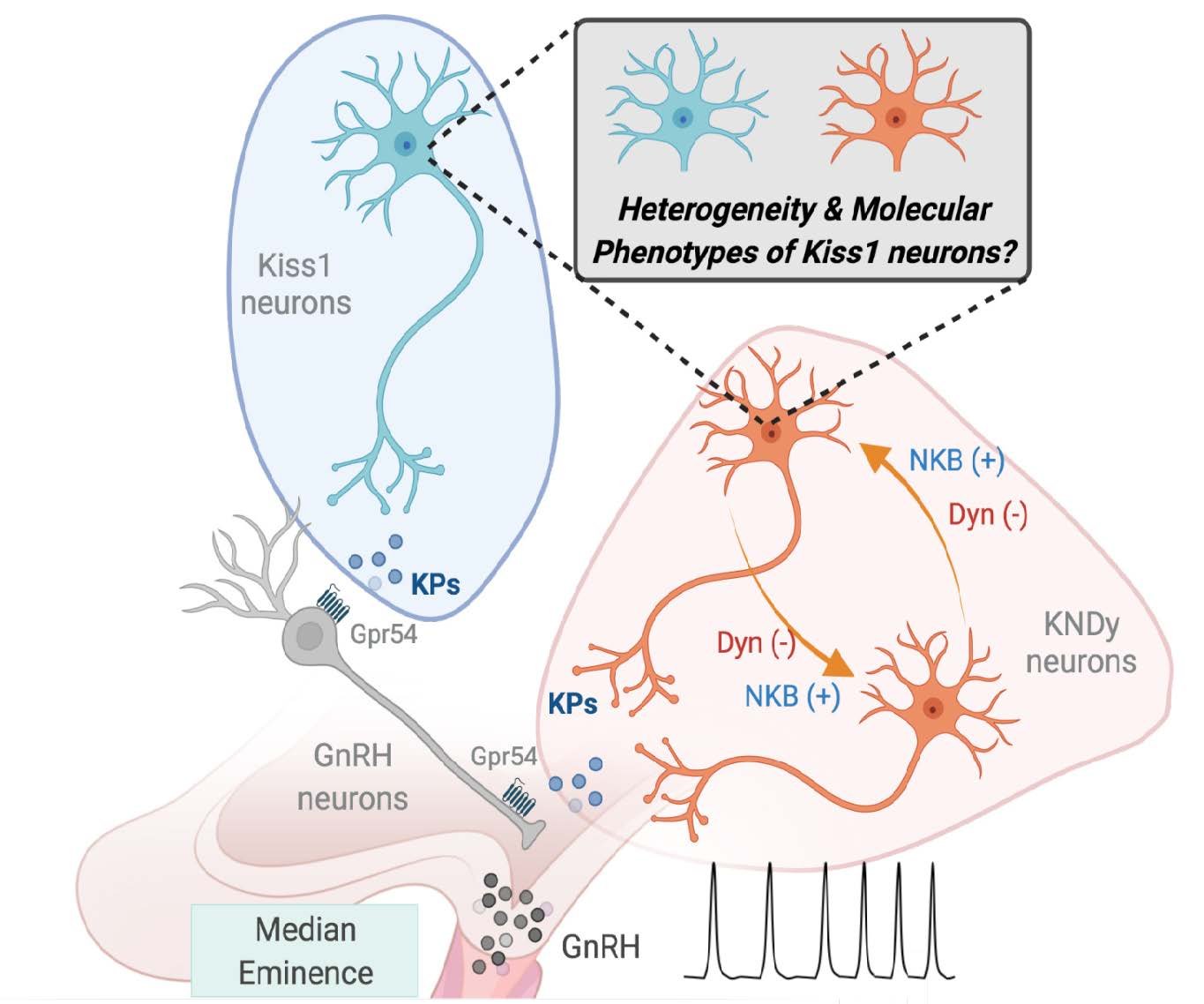
Characterisation of neuronal diversity
Define the molecular identity, transcriptional profiles, and functional heterogeneity of kisspeptin and dopaminergic neurons in the hypothalamus.
Elucidation of metabolic modulation
Investigate how nutrient and metabolic signals influence the activity and connectivity of these neuronal populations, with a focus on specific metabolic sensors and intracellular pathways.

Experimental validation of causal relationships
Apply targeted genome editing and functional manipulation to determine the mechanistic roles of identified pathways in regulating puberty onset and metabolic homeostasis.
Approach
DOPA-Kiss employs a multidisciplinary methodology that combines:
✅ Spatial and single-cell multi-omics to map metabolic and transcriptional landscapes.
✅ In vivo functional manipulation through DREADDs, optogenetics, and CRISPR/Cas9-mediated editing.
✅ Nutrient-sensing studies to identify metabolic determinants of pubertal regulation.
✅ Comparative analyses across sexes and models to ensure translational relevance.
By integrating these approaches, the project will bridge the gap between fundamental neuroendocrine research and the development of novel strategies to address puberty-related disorders.