Work Packages
An integrated framework combining multi-omics, functional neurobiology, and genome editing to dissect metabolic control of puberty.
Overview
The scientific strategy of DOPA-Kiss is articulated into three complementary and interconnected Work Packages (WP1–WP3). Each WP addresses a specific dimension of the project’s objectives, while maintaining strong integration to ensure coherent progress from molecular characterisation to functional validation and causal testing.
This structure enables a logical progression: from comprehensive identification of relevant molecular pathways (WP1), to mechanistic analysis (WP2), and finally to direct experimental demonstration of causality (WP3).
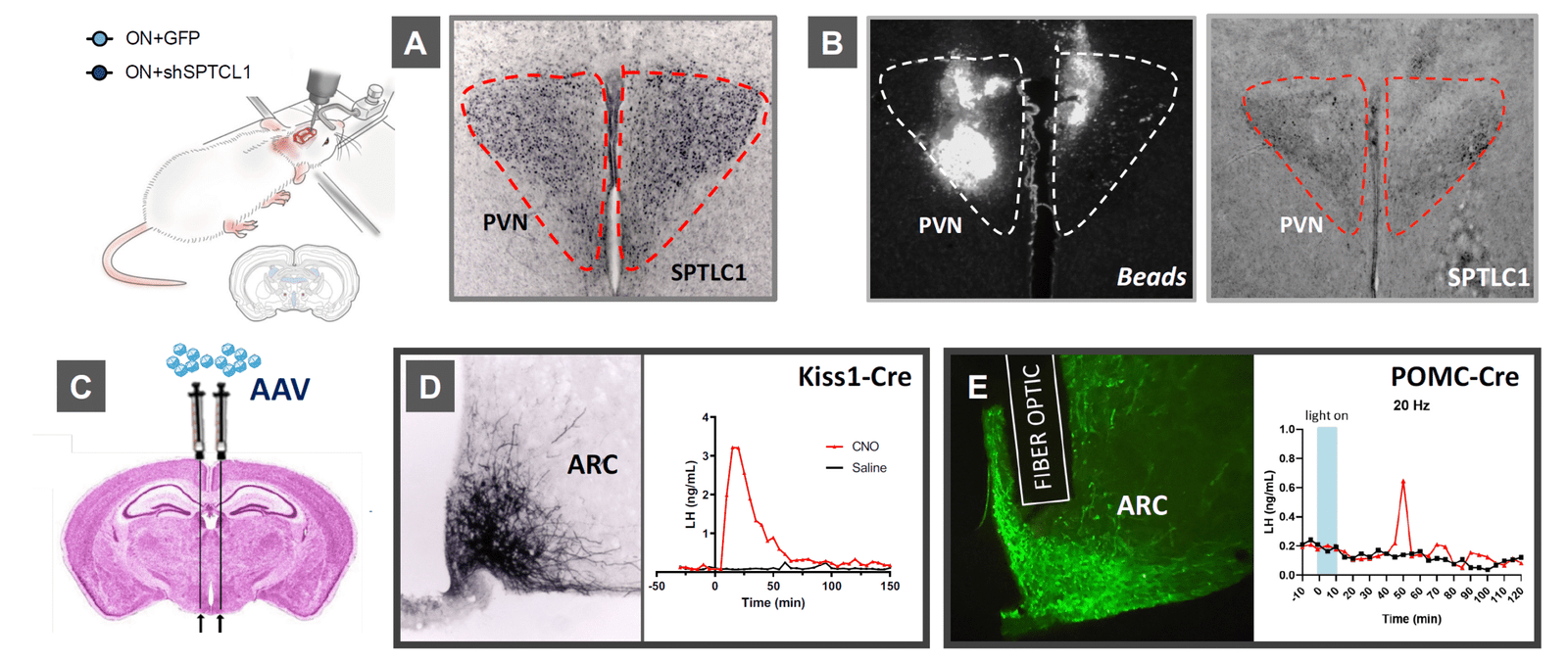
WP1
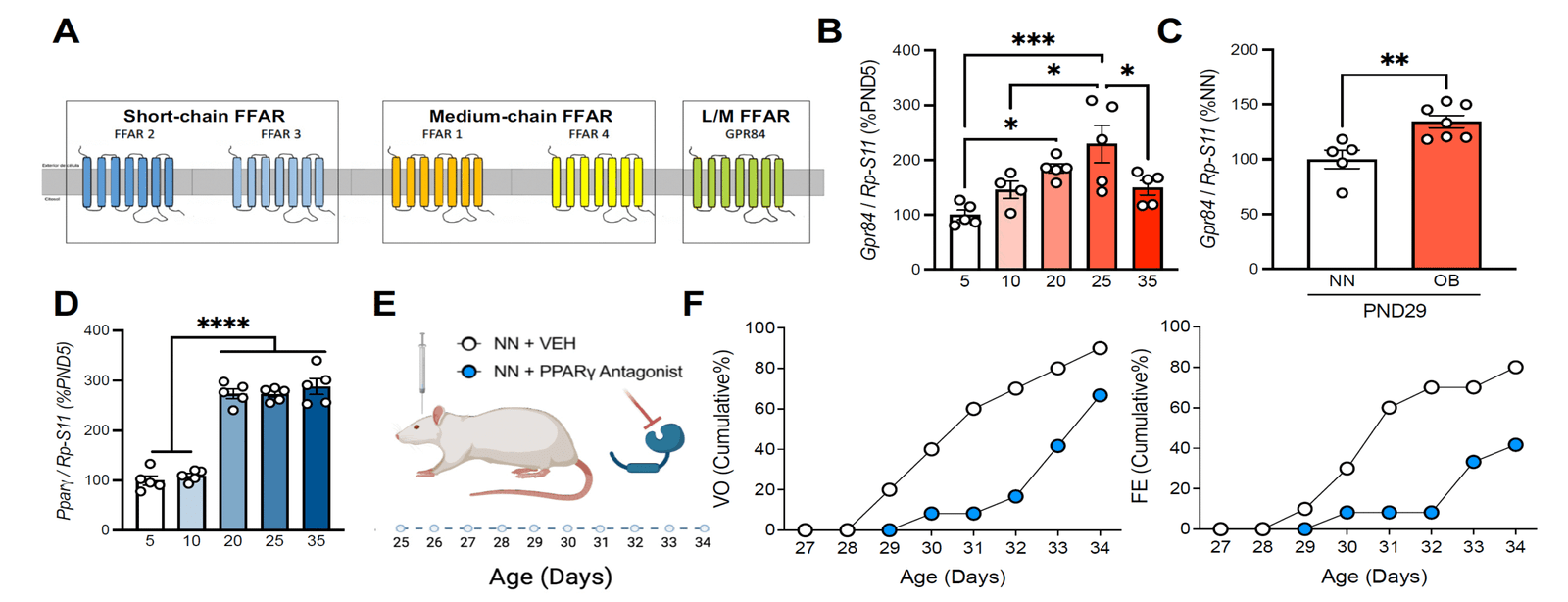
Multi-omics characterisation of hypothalamic kisspeptin and dopaminergic neurons
Aim:
To define the molecular, transcriptional, and metabolic profiles of hypothalamic kisspeptin and dopaminergic neurons under normal conditions and in models of early-onset obesity.
Key approaches:
✅ Comprehensive metabolomic and lipidomic profiling of the hypothalamus.
✅ Isolation of kisspeptin and dopaminergic neurons for pooled and single-cell transcriptomics and epigenomics.
Expected outcome:
A detailed molecular atlas identifying deregulated pathways and potential targets for functional testing in WP2 and WP3.
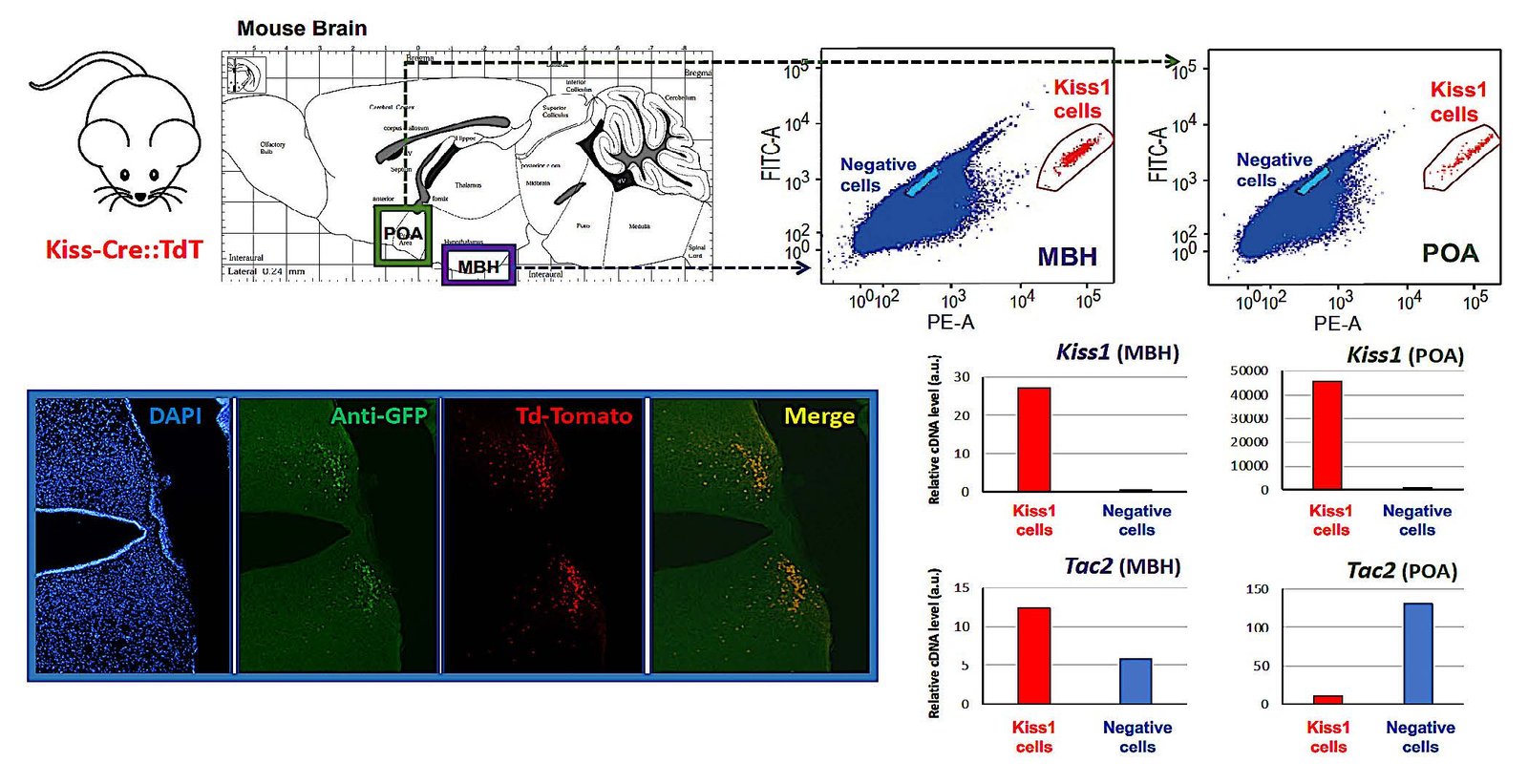
WP2
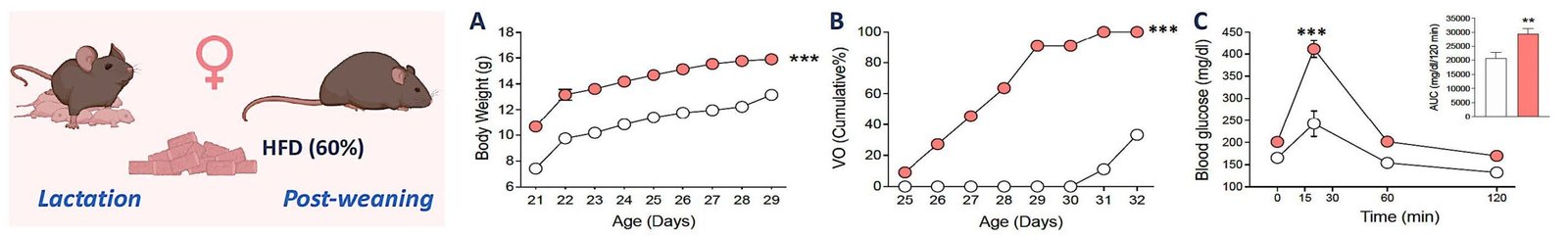
Mechanistic analysis of metabolic modulation
Aim:
To investigate how nutrient- and lipid-sensing pathways, stress responses, and autophagic mechanisms influence the function of kisspeptin and dopaminergic neurons.
Key approaches:
Expected outcome:
Identification of specific molecular mechanisms by which metabolic cues influence pubertal timing, including sex-specific effects.
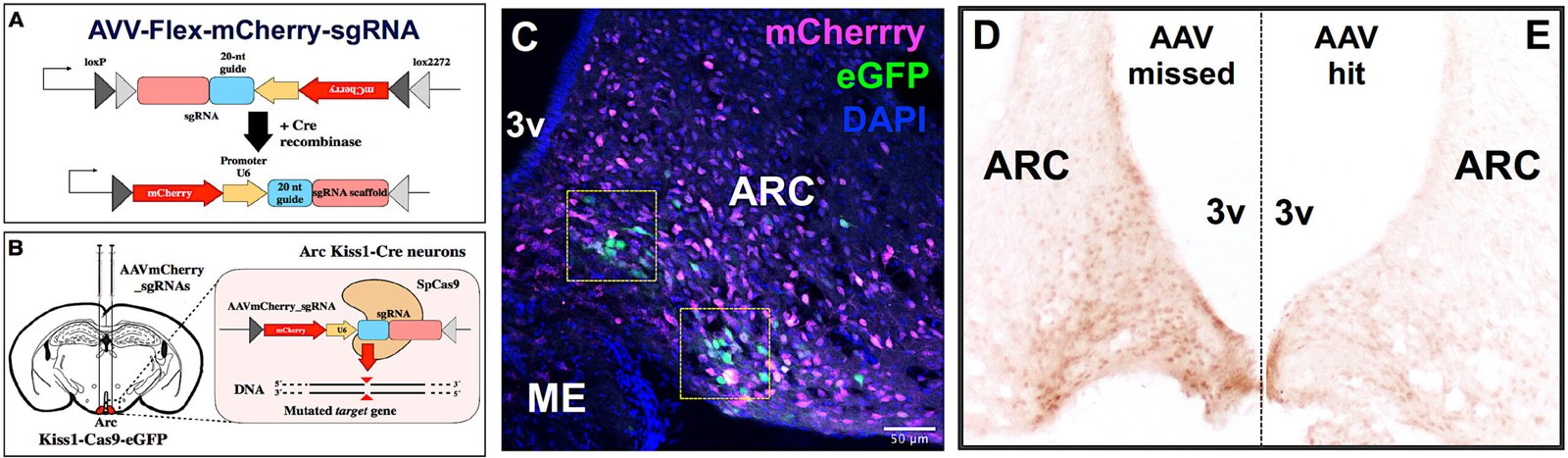
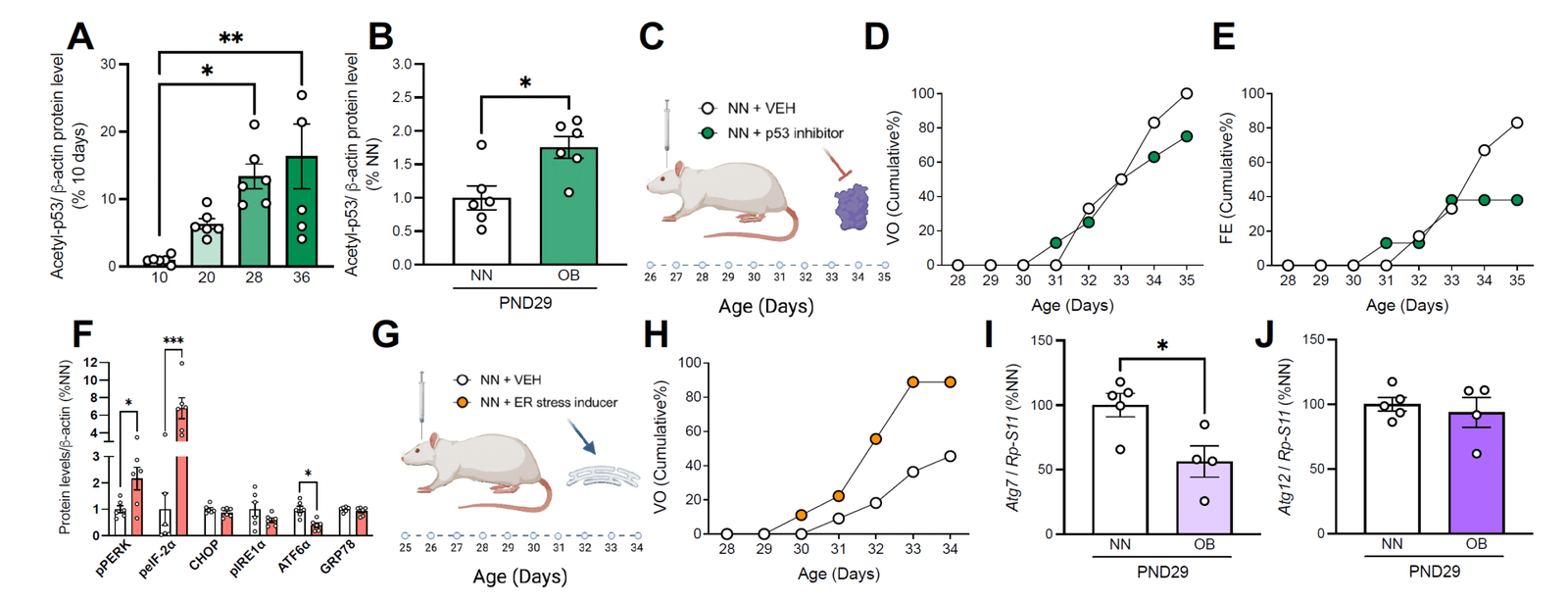
WP3
Causal validation through targeted genome editing
Aim:
To establish direct causal relationships between identified molecular pathways and the regulation of puberty and metabolism.
Key approaches:
Expected outcome:
Definitive experimental evidence linking specific molecular targets to the central control of puberty.